Detailed Report on Cloning Expression of Fermentation Gene of Interest
VerifiedAdded on 2023/04/23
|5
|1027
|480
Report
AI Summary
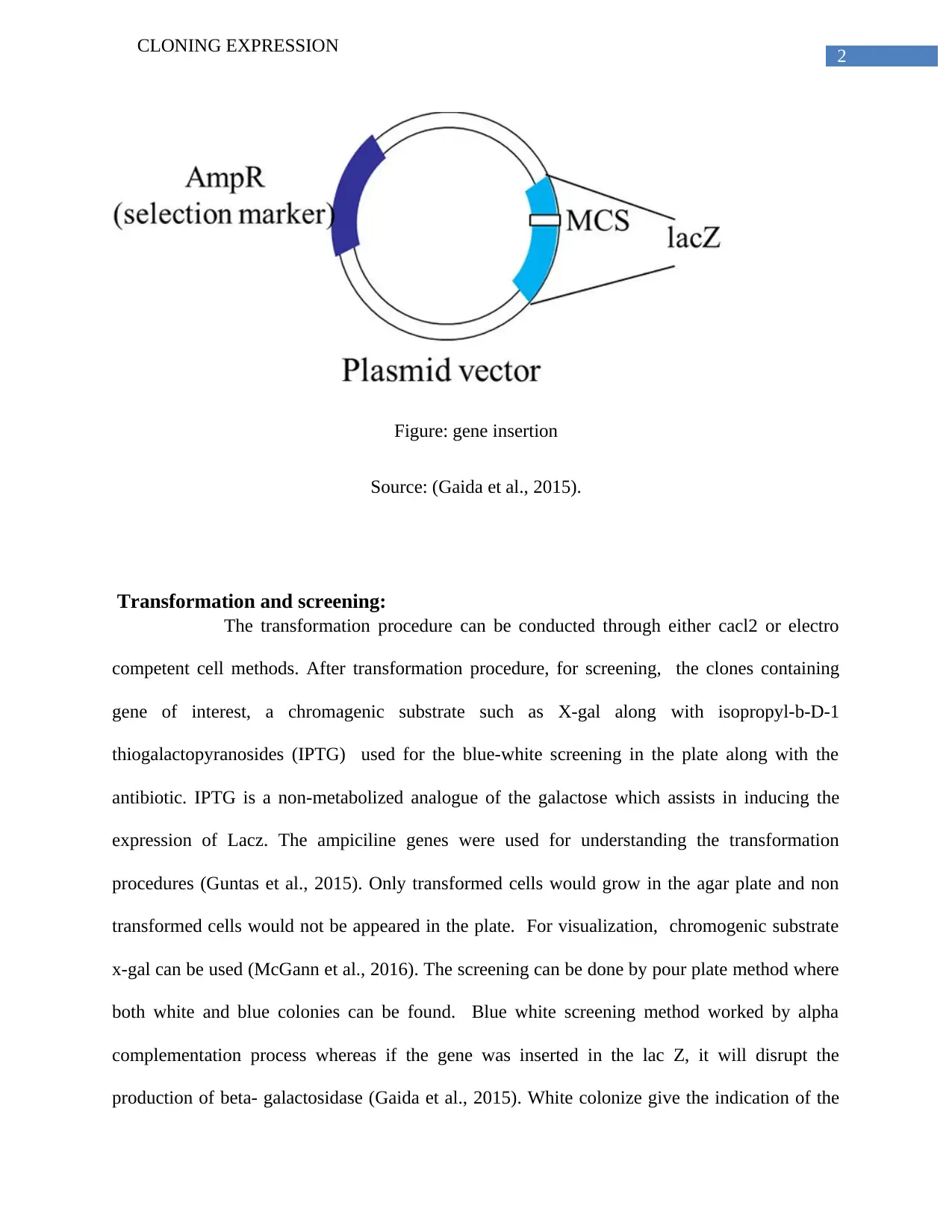
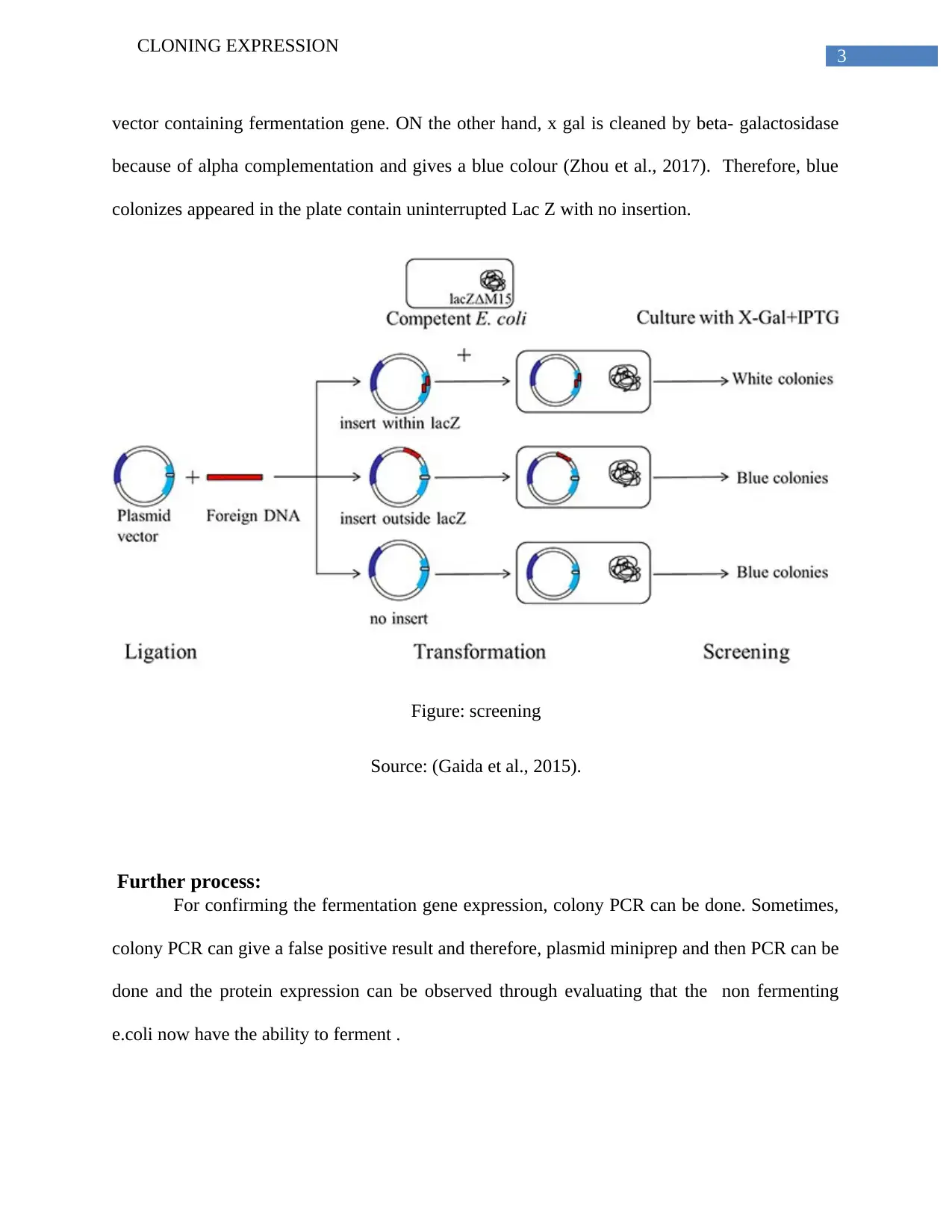
This report elucidates the process of cloning expression, focusing on the technique of using an expression vector to clone and express a specific protein, particularly a fermentation gene of interest. The report highlights blue-white screening as an effective method, enabled by alpha complementation, for achieving protein expression. It details the insertion of the gene, emphasizing the role of the lac Z operon and plasmid vectors like pBR322. The transformation process, using methods such as CaCl2 or electro-competent cells, is explained, followed by a description of screening techniques involving chromogenic substrates like X-gal and IPTG. The report further explains the use of antibiotic-resistant genes for understanding transformation procedures. Finally, the process of confirming fermentation gene expression through colony PCR and plasmid miniprep is discussed, noting potential false positives and the importance of verifying the fermentation ability of transformed E. coli.
1 out of 5








![[object Object]](/_next/static/media/star-bottom.7253800d.svg)