Protein Discovery and Purification of Bovine Chymosin: Analysis
VerifiedAdded on 2022/07/28
|11
|1519
|22
Report
AI Summary
This report provides a comprehensive analysis of bovine chymosin, an aspartyl protease extracted from the abomasum of suckling calves. It begins by detailing the primary structure of the protein, including its amino acid sequence, signal peptide, propeptide, and disulfide bonds. The report then explores the physico-chemical properties of chymosin, such as its molecular weight, theoretical pI, amino acid composition, and hydropathy plot, discussing how these properties influence its behavior and purification. The report further examines the initial and present purification processes of the protein, presenting flow charts that outline the key steps involved in separating and isolating chymosin from its source. The initial purification methods involved chromatographic columns and ion exchange, while the present purification methods involve filtration, precipitation, and centrifugation techniques to obtain purified bovine chymosin. Finally, the report includes a bibliography citing all primary sources used in the analysis.

Running head: PROTEIN DISCOVERY AND ANALYSIS
BOVINECHYMOSIN
Name of the Student
Name of the University
Author Note
BOVINECHYMOSIN
Name of the Student
Name of the University
Author Note
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1PROTEIN DISCOVERY AND ANALYSIS
Table of Contents
Introduction................................................................................................................................2
The primary structure of PET protein........................................................................................2
The physico-chemical properties of PET protein.......................................................................3
Initial purification of the protein................................................................................................5
Present purification of the protein..............................................................................................7
Bibliography...............................................................................................................................9
Table of Contents
Introduction................................................................................................................................2
The primary structure of PET protein........................................................................................2
The physico-chemical properties of PET protein.......................................................................3
Initial purification of the protein................................................................................................5
Present purification of the protein..............................................................................................7
Bibliography...............................................................................................................................9
2PROTEIN DISCOVERY AND ANALYSIS
Introduction
Bovine chymosin is an aspartyl protease extracted protein obtained from abomasum of
suckling calves. This protein is now synthesised in vivo as preprochymosin and has been
found to be secreted as prochymosin that is auto catalytically activated to form chymosin.
Chymosin has been found to be engineered inside laboratories and the gene encoding
chymosin has been cloned and expressed in bacteria and yeast also. This paper will discuss
the structure, physico-chemical properties and purification processes of bovine chymosin in
brief.
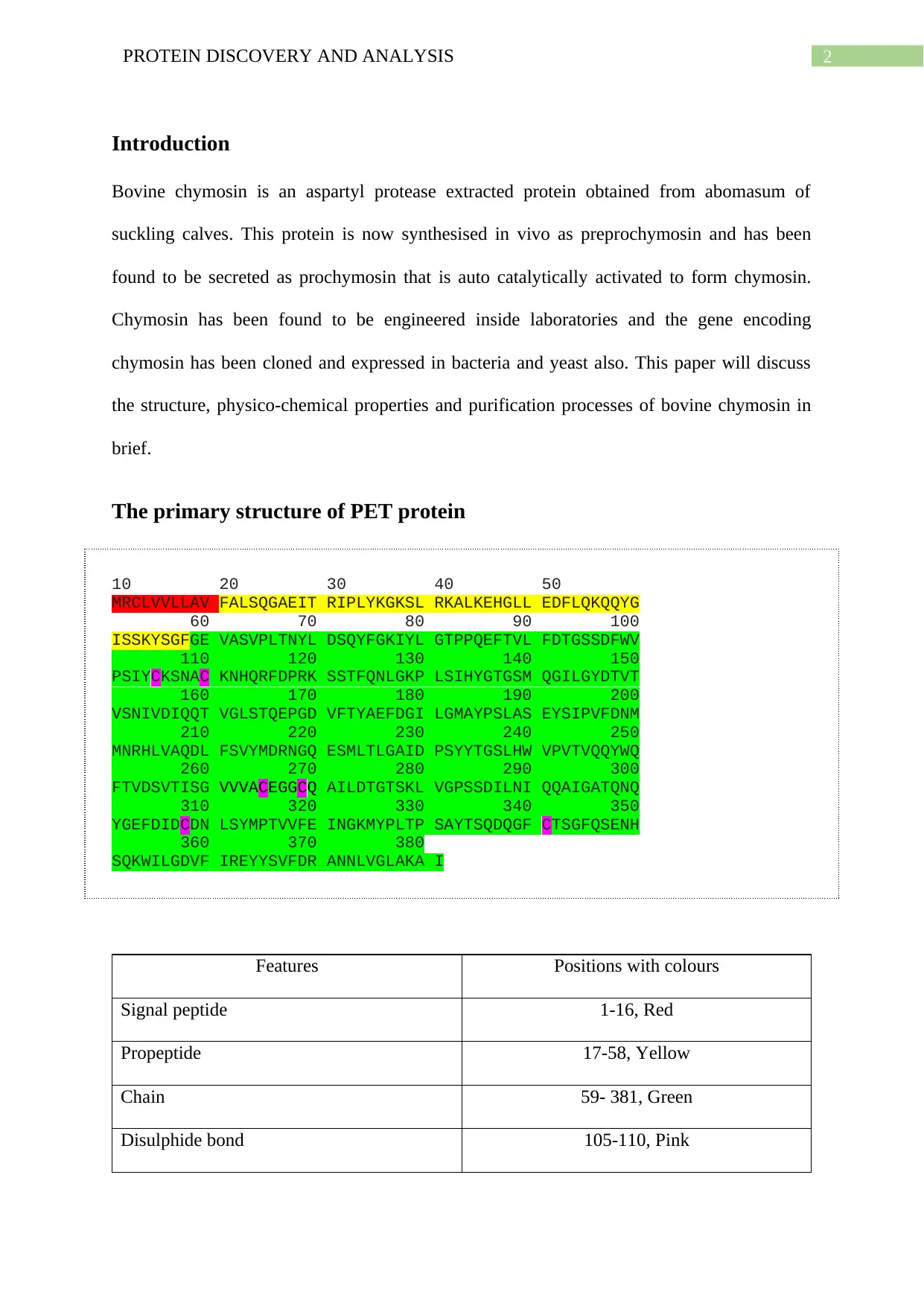
The primary structure of PET protein
10 20 30 40 50
MRCLVVLLAV FALSQGAEIT RIPLYKGKSL RKALKEHGLL EDFLQKQQYG
60 70 80 90 100
ISSKYSGFGE VASVPLTNYL DSQYFGKIYL GTPPQEFTVL FDTGSSDFWV
110 120 130 140 150
PSIYCKSNAC KNHQRFDPRK SSTFQNLGKP LSIHYGTGSM QGILGYDTVT
160 170 180 190 200
VSNIVDIQQT VGLSTQEPGD VFTYAEFDGI LGMAYPSLAS EYSIPVFDNM
210 220 230 240 250
MNRHLVAQDL FSVYMDRNGQ ESMLTLGAID PSYYTGSLHW VPVTVQQYWQ
260 270 280 290 300
FTVDSVTISG VVVACEGGCQ AILDTGTSKL VGPSSDILNI QQAIGATQNQ
310 320 330 340 350
YGEFDIDCDN LSYMPTVVFE INGKMYPLTP SAYTSQDQGF CTSGFQSENH
360 370 380
SQKWILGDVF IREYYSVFDR ANNLVGLAKA I
Features Positions with colours
Signal peptide 1-16, Red
Propeptide 17-58, Yellow
Chain 59- 381, Green
Disulphide bond 105-110, Pink
Introduction
Bovine chymosin is an aspartyl protease extracted protein obtained from abomasum of
suckling calves. This protein is now synthesised in vivo as preprochymosin and has been
found to be secreted as prochymosin that is auto catalytically activated to form chymosin.
Chymosin has been found to be engineered inside laboratories and the gene encoding
chymosin has been cloned and expressed in bacteria and yeast also. This paper will discuss
the structure, physico-chemical properties and purification processes of bovine chymosin in
brief.
The primary structure of PET protein
10 20 30 40 50
MRCLVVLLAV FALSQGAEIT RIPLYKGKSL RKALKEHGLL EDFLQKQQYG
60 70 80 90 100
ISSKYSGFGE VASVPLTNYL DSQYFGKIYL GTPPQEFTVL FDTGSSDFWV
110 120 130 140 150
PSIYCKSNAC KNHQRFDPRK SSTFQNLGKP LSIHYGTGSM QGILGYDTVT
160 170 180 190 200
VSNIVDIQQT VGLSTQEPGD VFTYAEFDGI LGMAYPSLAS EYSIPVFDNM
210 220 230 240 250
MNRHLVAQDL FSVYMDRNGQ ESMLTLGAID PSYYTGSLHW VPVTVQQYWQ
260 270 280 290 300
FTVDSVTISG VVVACEGGCQ AILDTGTSKL VGPSSDILNI QQAIGATQNQ
310 320 330 340 350
YGEFDIDCDN LSYMPTVVFE INGKMYPLTP SAYTSQDQGF CTSGFQSENH
360 370 380
SQKWILGDVF IREYYSVFDR ANNLVGLAKA I
Features Positions with colours
Signal peptide 1-16, Red
Propeptide 17-58, Yellow
Chain 59- 381, Green
Disulphide bond 105-110, Pink
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3PROTEIN DISCOVERY AND ANALYSIS
Disulphide bond 265-269, Pink
Disulphide bond 308-341, Pink
The physico-chemical properties of PET protein
Mature peptide sequence:
>sp|P00794|CHYM_BOVIN Chymosin OS=Bos taurus OX=9913 GN=CYM PE=1 SV=3
LLAVFALSQGAEITRIPLYKGKSLRKALKEHGLLEDFLQKQQYGISSKYSGFGE
VASVPLTNYLDSQYFGKIYLGTPPQEFTVLFDTGSSDFWVPSIYCKSNACKNHQRFDPRK
SSTFQNLGKPLSIHYGTGSMQGILGYDTVTVSNIVDIQQTVGLSTQEPGDVFTYAEFDGI
LGMAYPSLASEYSIPVFDNMMNRHLVAQDLFSVYMDRNGQESMLTLGAIDPSYYTGSLHW
VPVTVQQYWQFTVDSVTISGVVVACEGGCQAILDTGTSKLVGPSSDILNIQQAIGATQNQ
YGEFDIDCDNLSYMPTVVFEINGKMYPLTPSAYTSQDQGFCTSGFQSENHSQKWILGDVF
IREYYSVFDRANNLVGLAKAI
Physico-chemical properties:
User-provided sequence:
10 20 30 40 50 60
LLAVFALSQG AEITRIPLYK GKSLRKALKE HGLLEDFLQK QQYGISSKYS GFGEVASVPL
70 80 90 100 110 120
TNYLDSQYFG KIYLGTPPQE FTVLFDTGSS DFWVPSIYCK SNACKNHQRF DPRKSSTFQN
130 140 150 160 170 180
LGKPLSIHYG TGSMQGILGY DTVTVSNIVD IQQTVGLSTQ EPGDVFTYAE FDGILGMAYP
190 200 210 220 230 240
SLASEYSIPV FDNMMNRHLV AQDLFSVYMD RNGQESMLTL GAIDPSYYTG SLHWVPVTVQ
250 260 270 280 290 300
QYWQFTVDSV TISGVVVACE GGCQAILDTG TSKLVGPSSD ILNIQQAIGA TQNQYGEFDI
310 320 330 340 350 360
DCDNLSYMPT VVFEINGKMY PLTPSAYTSQ DQGFCTSGFQ SENHSQKWIL GDVFIREYYS
370
VFDRANNLVG LAKAI
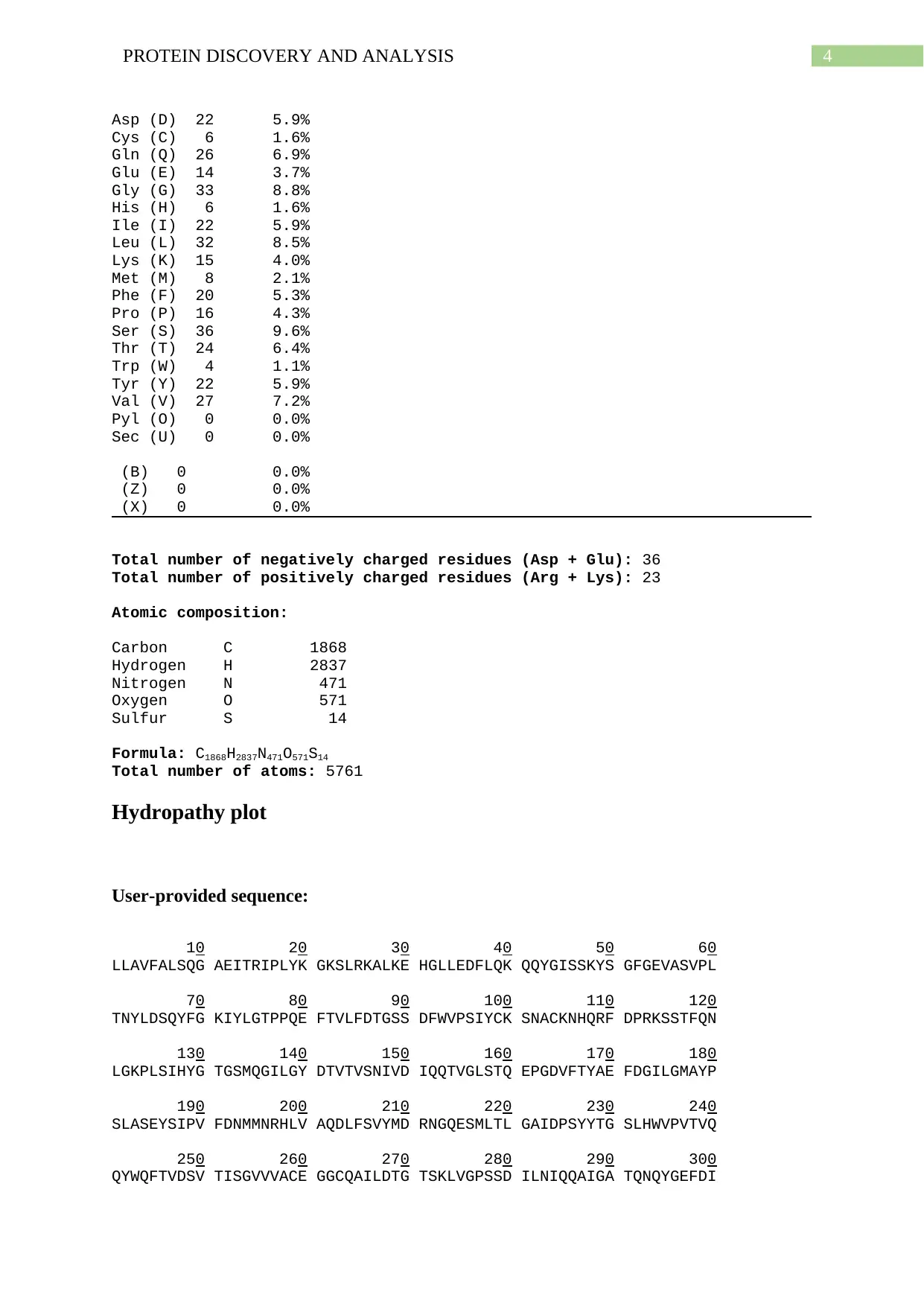
Number of amino acids: 375
Molecular weight: 41477.73
Theoretical pI: 4.82
Amino acid composition:
Ala (A) 19 5.1%
Arg (R) 8 2.1%
Asn (N) 15 4.0%
Disulphide bond 265-269, Pink
Disulphide bond 308-341, Pink
The physico-chemical properties of PET protein
Mature peptide sequence:
>sp|P00794|CHYM_BOVIN Chymosin OS=Bos taurus OX=9913 GN=CYM PE=1 SV=3
LLAVFALSQGAEITRIPLYKGKSLRKALKEHGLLEDFLQKQQYGISSKYSGFGE
VASVPLTNYLDSQYFGKIYLGTPPQEFTVLFDTGSSDFWVPSIYCKSNACKNHQRFDPRK
SSTFQNLGKPLSIHYGTGSMQGILGYDTVTVSNIVDIQQTVGLSTQEPGDVFTYAEFDGI
LGMAYPSLASEYSIPVFDNMMNRHLVAQDLFSVYMDRNGQESMLTLGAIDPSYYTGSLHW
VPVTVQQYWQFTVDSVTISGVVVACEGGCQAILDTGTSKLVGPSSDILNIQQAIGATQNQ
YGEFDIDCDNLSYMPTVVFEINGKMYPLTPSAYTSQDQGFCTSGFQSENHSQKWILGDVF
IREYYSVFDRANNLVGLAKAI
Physico-chemical properties:
User-provided sequence:
10 20 30 40 50 60
LLAVFALSQG AEITRIPLYK GKSLRKALKE HGLLEDFLQK QQYGISSKYS GFGEVASVPL
70 80 90 100 110 120
TNYLDSQYFG KIYLGTPPQE FTVLFDTGSS DFWVPSIYCK SNACKNHQRF DPRKSSTFQN
130 140 150 160 170 180
LGKPLSIHYG TGSMQGILGY DTVTVSNIVD IQQTVGLSTQ EPGDVFTYAE FDGILGMAYP
190 200 210 220 230 240
SLASEYSIPV FDNMMNRHLV AQDLFSVYMD RNGQESMLTL GAIDPSYYTG SLHWVPVTVQ
250 260 270 280 290 300
QYWQFTVDSV TISGVVVACE GGCQAILDTG TSKLVGPSSD ILNIQQAIGA TQNQYGEFDI
310 320 330 340 350 360
DCDNLSYMPT VVFEINGKMY PLTPSAYTSQ DQGFCTSGFQ SENHSQKWIL GDVFIREYYS
370
VFDRANNLVG LAKAI
Number of amino acids: 375
Molecular weight: 41477.73
Theoretical pI: 4.82
Amino acid composition:
Ala (A) 19 5.1%
Arg (R) 8 2.1%
Asn (N) 15 4.0%
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
4PROTEIN DISCOVERY AND ANALYSIS
Asp (D) 22 5.9%
Cys (C) 6 1.6%
Gln (Q) 26 6.9%
Glu (E) 14 3.7%
Gly (G) 33 8.8%
His (H) 6 1.6%
Ile (I) 22 5.9%
Leu (L) 32 8.5%
Lys (K) 15 4.0%
Met (M) 8 2.1%
Phe (F) 20 5.3%
Pro (P) 16 4.3%
Ser (S) 36 9.6%
Thr (T) 24 6.4%
Trp (W) 4 1.1%
Tyr (Y) 22 5.9%
Val (V) 27 7.2%
Pyl (O) 0 0.0%
Sec (U) 0 0.0%
(B) 0 0.0%
(Z) 0 0.0%
(X) 0 0.0%
Total number of negatively charged residues (Asp + Glu): 36
Total number of positively charged residues (Arg + Lys): 23
Atomic composition:
Carbon C 1868
Hydrogen H 2837
Nitrogen N 471
Oxygen O 571
Sulfur S 14
Formula: C1868H2837N471O571S14
Total number of atoms: 5761
Hydropathy plot
User-provided sequence:
10 20 30 40 50 60
LLAVFALSQG AEITRIPLYK GKSLRKALKE HGLLEDFLQK QQYGISSKYS GFGEVASVPL
70 80 90 100 110 120
TNYLDSQYFG KIYLGTPPQE FTVLFDTGSS DFWVPSIYCK SNACKNHQRF DPRKSSTFQN
130 140 150 160 170 180
LGKPLSIHYG TGSMQGILGY DTVTVSNIVD IQQTVGLSTQ EPGDVFTYAE FDGILGMAYP
190 200 210 220 230 240
SLASEYSIPV FDNMMNRHLV AQDLFSVYMD RNGQESMLTL GAIDPSYYTG SLHWVPVTVQ
250 260 270 280 290 300
QYWQFTVDSV TISGVVVACE GGCQAILDTG TSKLVGPSSD ILNIQQAIGA TQNQYGEFDI
Asp (D) 22 5.9%
Cys (C) 6 1.6%
Gln (Q) 26 6.9%
Glu (E) 14 3.7%
Gly (G) 33 8.8%
His (H) 6 1.6%
Ile (I) 22 5.9%
Leu (L) 32 8.5%
Lys (K) 15 4.0%
Met (M) 8 2.1%
Phe (F) 20 5.3%
Pro (P) 16 4.3%
Ser (S) 36 9.6%
Thr (T) 24 6.4%
Trp (W) 4 1.1%
Tyr (Y) 22 5.9%
Val (V) 27 7.2%
Pyl (O) 0 0.0%
Sec (U) 0 0.0%
(B) 0 0.0%
(Z) 0 0.0%
(X) 0 0.0%
Total number of negatively charged residues (Asp + Glu): 36
Total number of positively charged residues (Arg + Lys): 23
Atomic composition:
Carbon C 1868
Hydrogen H 2837
Nitrogen N 471
Oxygen O 571
Sulfur S 14
Formula: C1868H2837N471O571S14
Total number of atoms: 5761
Hydropathy plot
User-provided sequence:
10 20 30 40 50 60
LLAVFALSQG AEITRIPLYK GKSLRKALKE HGLLEDFLQK QQYGISSKYS GFGEVASVPL
70 80 90 100 110 120
TNYLDSQYFG KIYLGTPPQE FTVLFDTGSS DFWVPSIYCK SNACKNHQRF DPRKSSTFQN
130 140 150 160 170 180
LGKPLSIHYG TGSMQGILGY DTVTVSNIVD IQQTVGLSTQ EPGDVFTYAE FDGILGMAYP
190 200 210 220 230 240
SLASEYSIPV FDNMMNRHLV AQDLFSVYMD RNGQESMLTL GAIDPSYYTG SLHWVPVTVQ
250 260 270 280 290 300
QYWQFTVDSV TISGVVVACE GGCQAILDTG TSKLVGPSSD ILNIQQAIGA TQNQYGEFDI
5PROTEIN DISCOVERY AND ANALYSIS
310 320 330 340 350 360
DCDNLSYMPT VVFEINGKMY PLTPSAYTSQ DQGFCTSGFQ SENHSQKWIL GDVFIREYYS
370
VFDRANNLVG LAKAI
SEQUENCE LENGTH: 375
Using the scale Hphob. / Kyte & Doolittle, the individual values for the 20
amino acids are:
Ala: 1.800 Arg: -4.500 Asn: -3.500 Asp: -3.500 Cys: 2.500 Gln: -
3.500
Glu: -3.500 Gly: -0.400 His: -3.200 Ile: 4.500 Leu: 3.800 Lys: -
3.900
Met: 1.900 Phe: 2.800 Pro: -1.600 Ser: -0.800 Thr: -0.700 Trp: -
0.900
Tyr: -1.300 Val: 4.200 : -3.500 : -3.500 : -0.490
Weights for window positions 1,..,9, using linear weight variation model:
1 2 3 4 5 6 7 8 9
1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00
edge center edge
310 320 330 340 350 360
DCDNLSYMPT VVFEINGKMY PLTPSAYTSQ DQGFCTSGFQ SENHSQKWIL GDVFIREYYS
370
VFDRANNLVG LAKAI
SEQUENCE LENGTH: 375
Using the scale Hphob. / Kyte & Doolittle, the individual values for the 20
amino acids are:
Ala: 1.800 Arg: -4.500 Asn: -3.500 Asp: -3.500 Cys: 2.500 Gln: -
3.500
Glu: -3.500 Gly: -0.400 His: -3.200 Ile: 4.500 Leu: 3.800 Lys: -
3.900
Met: 1.900 Phe: 2.800 Pro: -1.600 Ser: -0.800 Thr: -0.700 Trp: -
0.900
Tyr: -1.300 Val: 4.200 : -3.500 : -3.500 : -0.490
Weights for window positions 1,..,9, using linear weight variation model:
1 2 3 4 5 6 7 8 9
1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00
edge center edge
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6PROTEIN DISCOVERY AND ANALYSIS
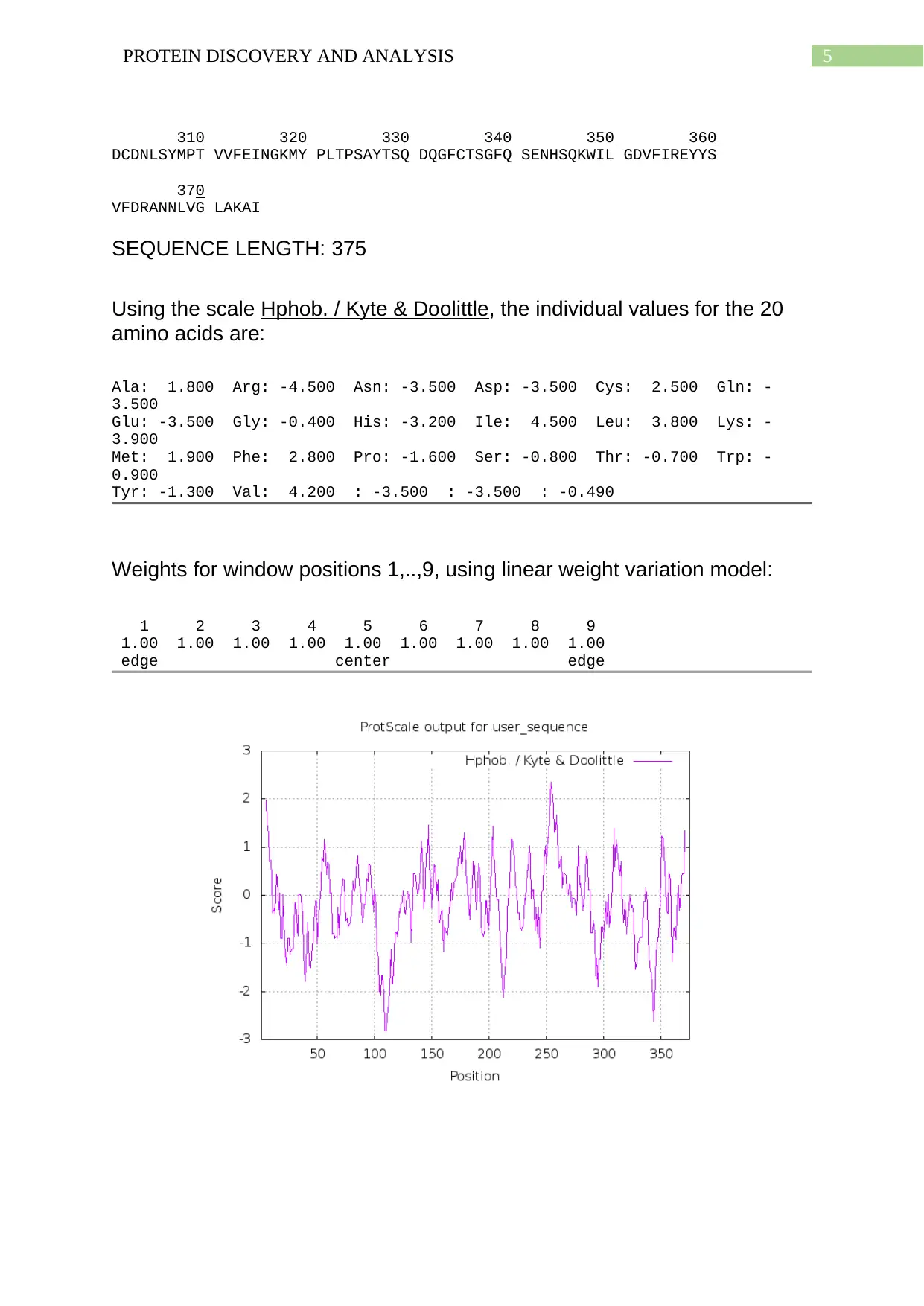
A hydropathy plot is a graphical plot that allows the visualisation of hydrophobicity and
hydrophilicity over the complete peptide length sequence. The hydropathy scale has been
found to be based on the hydrophilic and hydrophobic properties of all the 20 amino acids
which have been found to be the main composition of the protein used in this report (Johri et
al., 2019). The is a positively charged amino acid dominated protein as observed from the
amino acid composition. The highest peak at the positive quadrant has been shown at the
position 260 which states that the amino acid is well inside the core of the protein. Thus, it
can be stated that this amino acid is hydrophobic in nature. The highest peak at the negative
quadrant has been found at 120 position. This proves that the amino acid is will outside the
protein, in contact with the surrounding environment with water. Thus, it can be stated that
this amino acid is hydrophilic in nature.
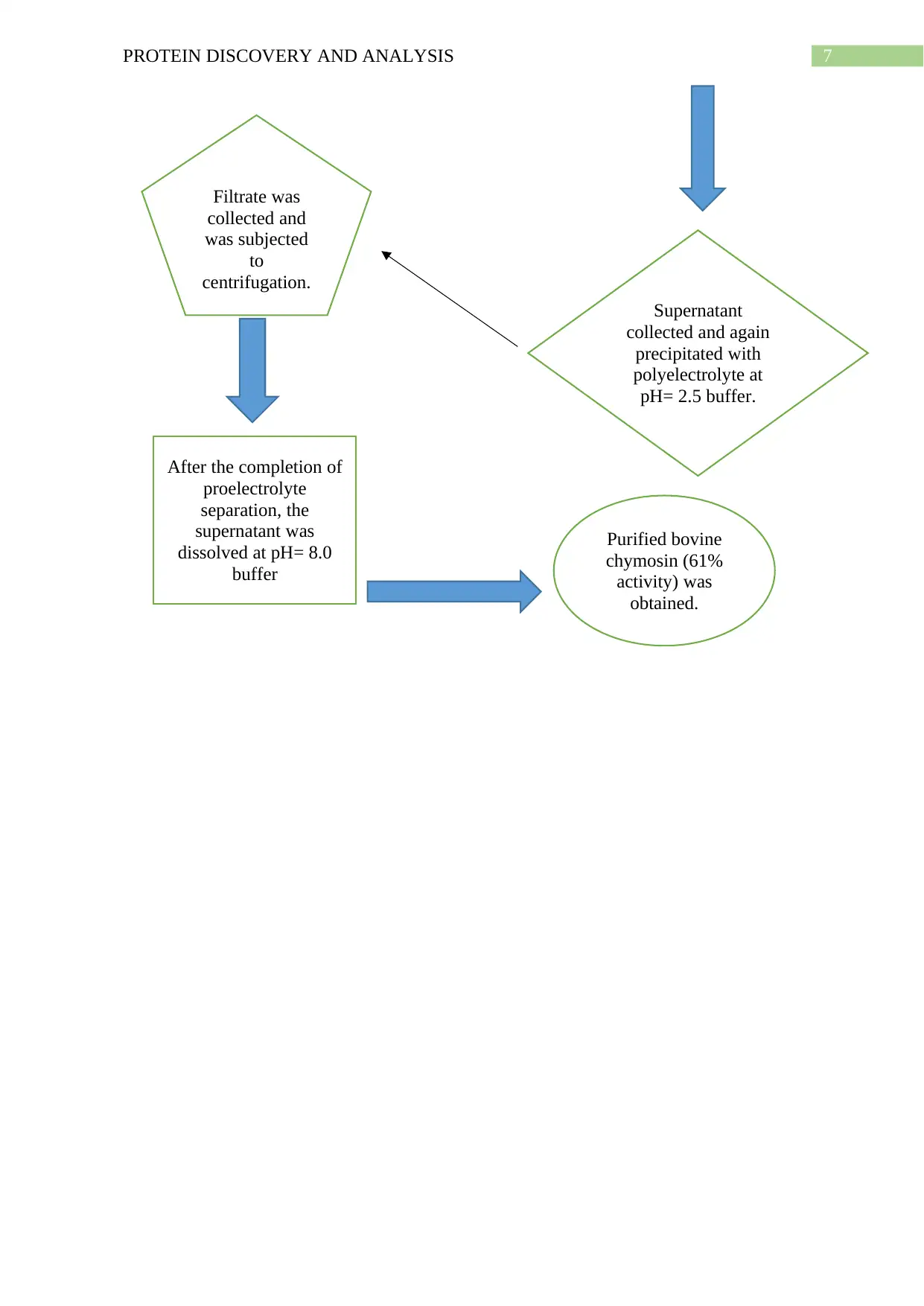
Present purification of the protein
The flow chart of the process has been given below-
Bovine
chymosin
collected Filtered and
filtrate was
collected-
filtrate
The collected
filtrate was
precipitated with
negatively
charged
electrolyte Zymogen
was
activated
after
adding
trypsin
A hydropathy plot is a graphical plot that allows the visualisation of hydrophobicity and
hydrophilicity over the complete peptide length sequence. The hydropathy scale has been
found to be based on the hydrophilic and hydrophobic properties of all the 20 amino acids
which have been found to be the main composition of the protein used in this report (Johri et
al., 2019). The is a positively charged amino acid dominated protein as observed from the
amino acid composition. The highest peak at the positive quadrant has been shown at the
position 260 which states that the amino acid is well inside the core of the protein. Thus, it
can be stated that this amino acid is hydrophobic in nature. The highest peak at the negative
quadrant has been found at 120 position. This proves that the amino acid is will outside the
protein, in contact with the surrounding environment with water. Thus, it can be stated that
this amino acid is hydrophilic in nature.
Present purification of the protein
The flow chart of the process has been given below-
Bovine
chymosin
collected Filtered and
filtrate was
collected-
filtrate
The collected
filtrate was
precipitated with
negatively
charged
electrolyte Zymogen
was
activated
after
adding
trypsin
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
7PROTEIN DISCOVERY AND ANALYSIS
Supernatant
collected and again
precipitated with
polyelectrolyte at
pH= 2.5 buffer.
Filtrate was
collected and
was subjected
to
centrifugation.
After the completion of
proelectrolyte
separation, the
supernatant was
dissolved at pH= 8.0
buffer
Purified bovine
chymosin (61%
activity) was
obtained.
Supernatant
collected and again
precipitated with
polyelectrolyte at
pH= 2.5 buffer.
Filtrate was
collected and
was subjected
to
centrifugation.
After the completion of
proelectrolyte
separation, the
supernatant was
dissolved at pH= 8.0
buffer
Purified bovine
chymosin (61%
activity) was
obtained.
8PROTEIN DISCOVERY AND ANALYSIS
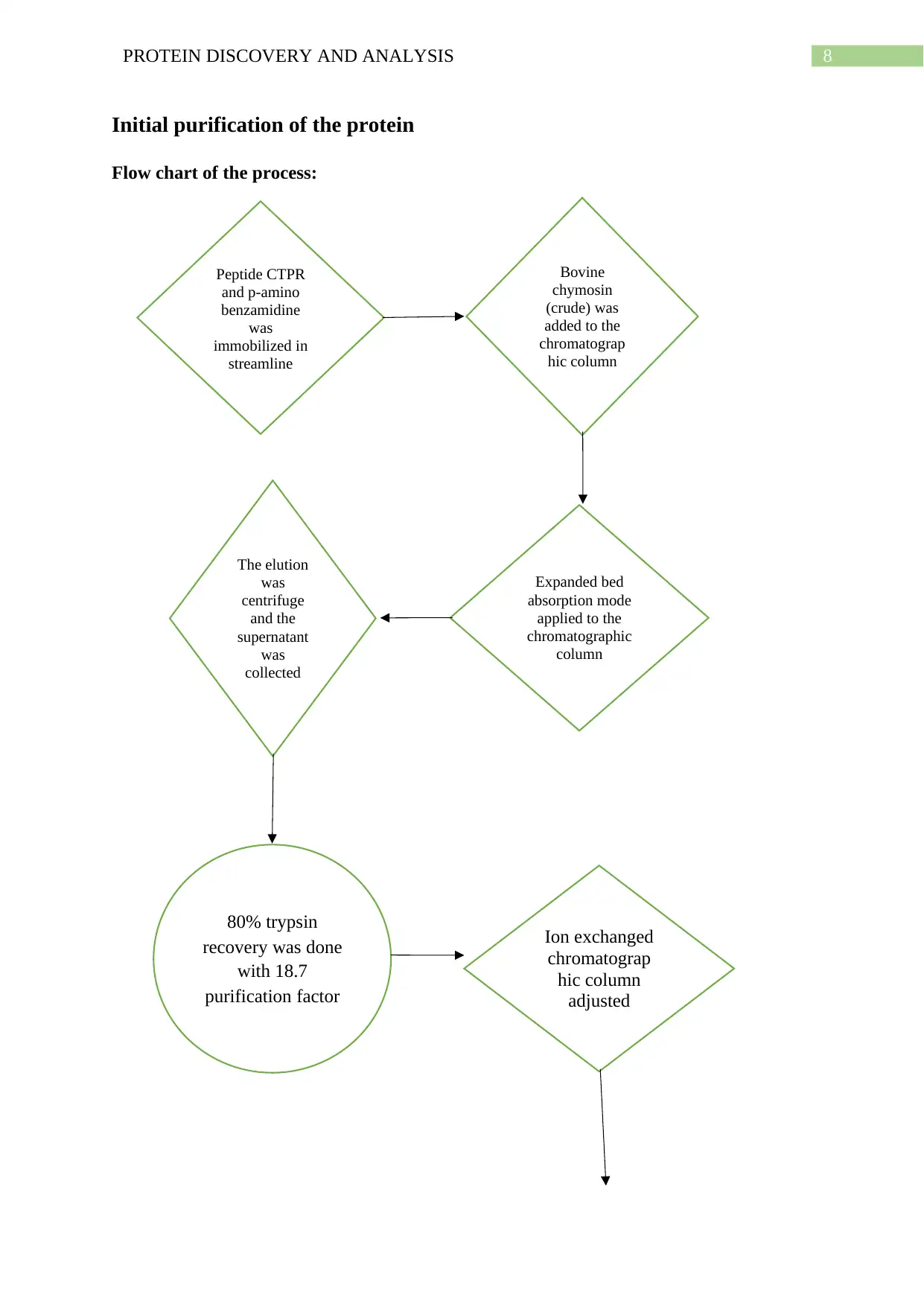
Initial purification of the protein
Flow chart of the process:
Peptide CTPR
and p-amino
benzamidine
was
immobilized in
streamline
Bovine
chymosin
(crude) was
added to the
chromatograp
hic column
Expanded bed
absorption mode
applied to the
chromatographic
column
The elution
was
centrifuge
and the
supernatant
was
collected
80% trypsin
recovery was done
with 18.7
purification factor
Ion exchanged
chromatograp
hic column
adjusted
Initial purification of the protein
Flow chart of the process:
Peptide CTPR
and p-amino
benzamidine
was
immobilized in
streamline
Bovine
chymosin
(crude) was
added to the
chromatograp
hic column
Expanded bed
absorption mode
applied to the
chromatographic
column
The elution
was
centrifuge
and the
supernatant
was
collected
80% trypsin
recovery was done
with 18.7
purification factor
Ion exchanged
chromatograp
hic column
adjusted
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
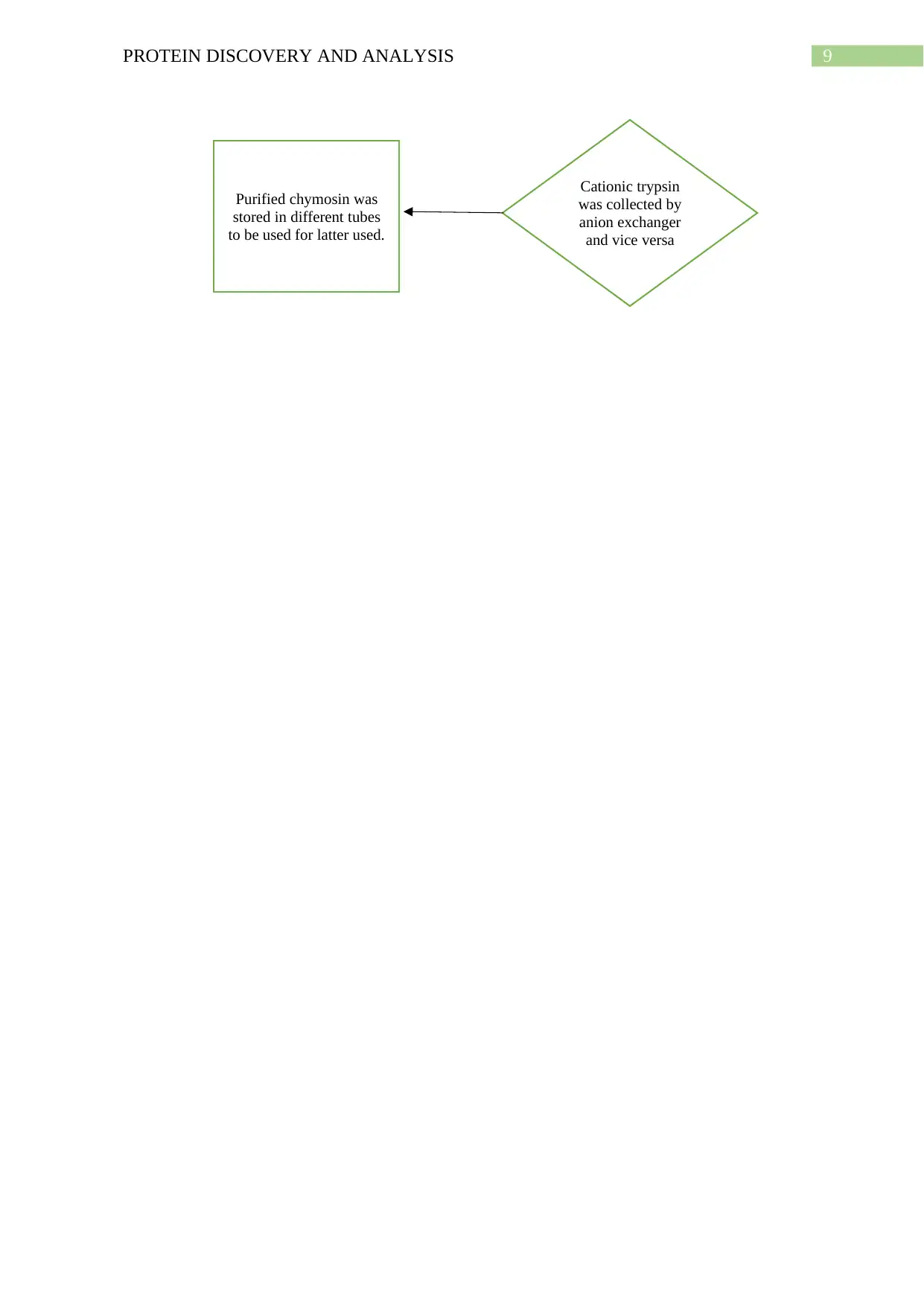
9PROTEIN DISCOVERY AND ANALYSIS
Cationic trypsin
was collected by
anion exchanger
and vice versa
Purified chymosin was
stored in different tubes
to be used for latter used.
Cationic trypsin
was collected by
anion exchanger
and vice versa
Purified chymosin was
stored in different tubes
to be used for latter used.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

10PROTEIN DISCOVERY AND ANALYSIS
Bibliography
Bárbara, B., Emilia, B. M., Gastón, K., Guillermo, P., & Nadia, W. V. (2020). Design and
optimization of an alternative chymosin purification method by adsorption onto non-
soluble alginate–carrageenan bed. Polymer Bulletin, 1-20.
Boeris, V., Romanini, D., Farruggia, B., & Picó, G. (2009). Purification of chymosin from
bovine pancreas using precipitation with a strong anionic polyelectrolyte. Process
Biochemistry, 44(5), 588-592.
Egelrud, T. (1993). Purification and preliminary characterization of stratum corneum
chymotryptic enzyme: a proteinase that may be involved in desquamation. Journal of
investigative dermatology, 101(2), 200-204.
Ruan, J., Yan, J., Chen, H., Jianping, C., Sun, W., & Zhao, G. (2017). Purification and
properties of the chymosin inhibitor from wild emmer wheat (Triticum dicoccoides)
of Israel and its toxic effect on beet armyworm, Spodoptera exigua. Pesticide
biochemistry and physiology, 142, 141-147.
ProtScale
ExPASy
NCBI
Bibliography
Bárbara, B., Emilia, B. M., Gastón, K., Guillermo, P., & Nadia, W. V. (2020). Design and
optimization of an alternative chymosin purification method by adsorption onto non-
soluble alginate–carrageenan bed. Polymer Bulletin, 1-20.
Boeris, V., Romanini, D., Farruggia, B., & Picó, G. (2009). Purification of chymosin from
bovine pancreas using precipitation with a strong anionic polyelectrolyte. Process
Biochemistry, 44(5), 588-592.
Egelrud, T. (1993). Purification and preliminary characterization of stratum corneum
chymotryptic enzyme: a proteinase that may be involved in desquamation. Journal of
investigative dermatology, 101(2), 200-204.
Ruan, J., Yan, J., Chen, H., Jianping, C., Sun, W., & Zhao, G. (2017). Purification and
properties of the chymosin inhibitor from wild emmer wheat (Triticum dicoccoides)
of Israel and its toxic effect on beet armyworm, Spodoptera exigua. Pesticide
biochemistry and physiology, 142, 141-147.
ProtScale
ExPASy
NCBI
1 out of 11
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.




